sickle cell anemia has been traced to what type of mutation?
Abstract
Genetic analysis of admixed populations raises special concerns with regard to study design and data processing, especially to avoid population stratification biases. The bespeak mutation responsible for sickle prison cell anaemia codes for a variant hemoglobin, sickle hemoglobin or HbS, whose presence drives the pathophysiology of illness. Here we propose to explore ancestry and population construction in a genome-broad study with particular accent on chromosome xi in ii SCA admixed cohorts obtained from urban populations of Brazil (Pernambuco and São Paulo) and the U.s. (Pennsylvania). Ancestry inference showed different proportions of European, African and American backgrounds in the composition of our samples. Brazilians were more than admixed, had a lower African background (43% vs. 78% on the genomic level and 44% vs. 76% on chromosome 11) and presented a signature of positive selection and Iberian introgression in the HbS region, driving a high differentiation of this locus between the two cohorts. The genetic structures of the SCA cohorts from Brazil and US differ considerably on the genome-broad, chromosome xi and HbS mutation locus levels.
Introduction
Sickle cell anaemia (SCA) is caused past homozygosity for a point mutation in the beta-globin gene (HBB) on chromosome 11. SCA was the first monogenic disease to be described in humans1 and manifestations are caused by red blood cells damaged by HbS2. Five RFLP-assessed haplotypes, named afterwards the locations where they occur more frequently (Benin, Central Africa Republic or Car, Cameroon, Senegal and Arab-Indian), are classically used to classify the HBB cluster. High fetal haemoglobin (HbF) levels are associated with the Senegal and Arab-Indian haplotypes, compared to the Benin, CAR and Cameroon haplotypes3. Individuals with CAR haplotypes tend to nowadays the lowest HbF levels, while individuals with the Republic of benin haplotype usually have intermediate HbF product levelsfour. Despite the fact that the protective effect of HbF may vary according to its distribution amongst erythrocytes, every bit shown by severe SCA cases conveying the Arab-Indian haploytpe5, these findings have motivated arable label of diverse SCA populations worldwide regarding HBB haplotypes.
Some effort has been made to draw genetic diversity and structure among SCA patientshalf-dozen,7,eight,nine. Even so, aspects regarding the upshot of European beginnings10 and fine genetic structure on the SCA mutation locus remain elusive. Large association studies accept been mostly conducted on SCA patients from the US (SUS). Other studies, such as those conducted in Brazilian SCA patients (SBR), rely frequently on findings from studies of the SUS population. The United states of america and Brazil take the highest prevalence of new-borns with SCA on the American continent, estimated to be iv,351 and 2,978, respectively, in 201011 and are divergent in demographic history regarding migration and admixture.
Here, we aim to clarify how admixed populations affected by SCA diverge from each other. Also, we aim to further describe the beginnings of SCA patients from Brazil, who have been explored relatively fiddling compared to U.s.a. patients. To achieve these goals, nosotros propose to compare genetic structures of two populations at the genome-broad and local levels, through the assay of a Due north American cohort (from the Children's Hospital of Philadelphia-PA) and a Brazilian SCA cohort (from the Haematology-Haemotherapy Centre in Campinas-SP, HEMOCENTRO, and Haematology and Haemotherapy Foundation of Pernambuco-PE, HEMOPE), using high-density genome-broad microarrays (Genome-Wide Human being SNP Array 6.0, Affymetrix Inc., CA, U.s.).
Results
We evaluated the genetic structure at the genome-wide and chromosomal level by comparing SCA cohorts from the U.s. and Brazil, along with nineteen worldwide populations from the African, European, American and Asian continents. Unaffected people (HbAA genotype) sampled from The states and Brazil (AAM and BRZ, respectively), were also included (see Supplementary Table S1).
Genomic data from all populations were analysed by keeping 155,820 SNPs after quality control for a principal component assay (PCA), depicted in Supplementary Fig. S1. As expected, BRZ genetic variation is displayed as very heterogeneous in the PCA, with individuals being dispersed between European and African populations, via a pattern demonstrated earlier12,13. We also computed Hudson'southward fixation index (FST) as a measure of genetic distance between populations (Supplementary Table S2) and constitute SBR to exist closer to Europeans, relatively to SUS. PCA and FST were likewise concordant in depicting SCA patients from the United states of america and Brazil as closer to each other (FST = 0.017) than to Europeans (values ranging from 0.03 to 0.088).
By estimating mean ancestries for each one of the 23 populations (Fig. one and Table one), we establish that both European and African ancestries are predominant in the afflicted cohorts. Table 1 depicts mean ancestries for the SCA groups by geographical region. The South European (presumably Iberian and Italian) ancestral component estimate is more than prominent in Brazilians (both affected and non-affected). Eastern African (Bantu) corresponds to a larger proportion of within-Africa ancestry in Brazilians relatively to US samples' hateful estimates. BRZ was close to SBR, except that the latter seems to take slightly more African ancestry, while SUS individuals seem to take a somewhat lower mean African component compared to AAM (Fig. 1). On a global scale, the mean beginnings of US patients is 78% African, 19% European and 1.5% Amerindian, while affected Brazilians testify a mean ancestry of 43%, 45% and 10%, respectively, consistent with previous reports8,9. Moreover, African components are divided into 38.iii% Western-Africa Mandé-related and 39.four% Eastern Bantu-related on average for the SUS, while SBR present fifteen.three% Mandé-related and 28% Bantu-related ancestries (Table 1).
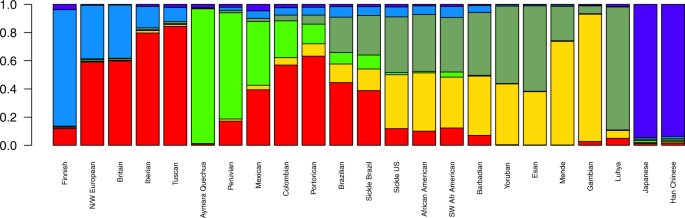
Mean ancestral components inferred by ADMIXTURE analysis. This analysis was performed using 155,820 SNPs across the genome. M = 6 had the everyman cantankerous-validation error and thus was selected to correspond bequeathed components. Each bar represents a population in x-axis, while y-axis depicts mean proportional ancestry for each population (run across Supplementary Table S1 for details on each population). N/W: Due north and West; SW: Southwest; Sickle: sickle cell anaemia.
We besides explored ancestries on chromosome 11, where the HBB factor cluster is located (Fig. 2, Tables ane and 2 and Supplementary Figs S2 and S3). In contrast to Brazilians, the affected American cohort typically shows more than 70% of African haplotypes along the chromosome. SBR had a more balanced constitution, showing 44% African and 39.iii% European haplotypes inferred from the phased data on boilerplate (while SUS had an estimated mean of 18.3% of haplotypes of European origin), run across Tabular array 1. SCA cohorts also diverged in supposedly Native American proportions (mean five.7% vs. 16.7% for SUS and SBR, respectively). Of note, the ii populations evaluated at the chromosomal level had a like tiptop, evidencing predominance of African haplotypes on 11p15.four, where the HBB region is located (Fig. 2a), except for a 1.ii Mb region where African ancestry estimates drop sharply for Brazilians.
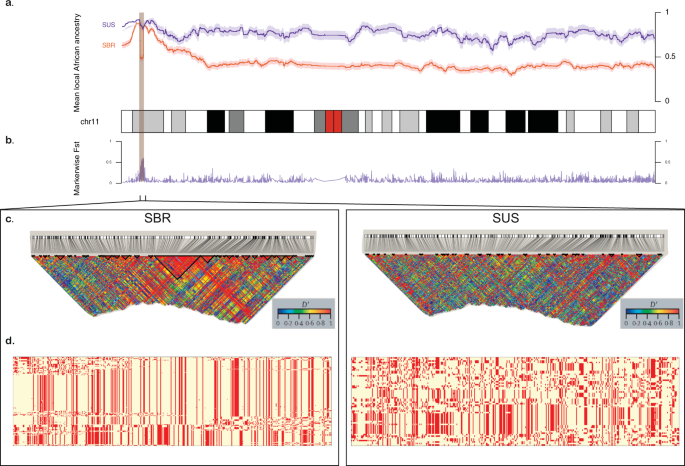
Comparing between Brazilian (SBR) and American (SUS) sickle cell anaemia patients on chromosome 11. (a) Diagram of the chromosome 11 (27,188 SNPs). Higher panel: x-axis represents physical position, y-centrality is local mean African component inferred by SABER+; shades denote standard errors. (b) FST values for each marking showing high differentiation on the HBB cluster region (highlighted), also a site where SBR shows a drib in mean African beginnings. (c) Linkage disequilibrium in Golden heat map generated by Haploview for SBR (left) and SUS (right) cohorts. (d) Phased haplotypes diagram along the highlighted expanse (chromosome 11:4.5–five.7 Mb) for SBR (left) and SUS (right).
We besides found SUS and SBR to accept highly divergent allele frequencies in a region at 11p15.4, as highlighted in Fig. 2b, measured by marking-wise Weir and Cockerham's FST estimates. Additionally, markedly different LD and haplotype structures (Fig. 2c,d) were establish in the same region. SBR subjects have a well-defined 266 kb LD block comprising the HBB cluster and a office of the locus control region (LCR), while SUS subjects take a xiii kb block upstream of the cluster and scattered regions of high LD (Fig. 2c). The conventional classification by RFLP-defined haplotypes was inferred in silico and conformed to expected proportions for both SCA cohorts (Tabular array 2). Nosotros next conducted the integrated haplotype score (iHS), and found a region in which Brazilians have markers with iHS values ranging from −3.8 to −four.5 (Fig. 3a), indicating that in that location are alleles showing a design of extended haplotype homozygosity (EHH), probably a effect of recent selective sweep. To test if this signal is replicated in SUS patients, we conducted a cantankerous-population EHH calculation (XP-EHH, Fig. 3b) and found this measure to converge towards a value of 2, suggesting that a recent positive selection event took place on the SBR population nigh the chr11:5.iv–5.5 Mb region, merely not in the SUS, consistent with the abovementioned difference in FST values (also depicted for this region, dashed line in Fig. 3c).
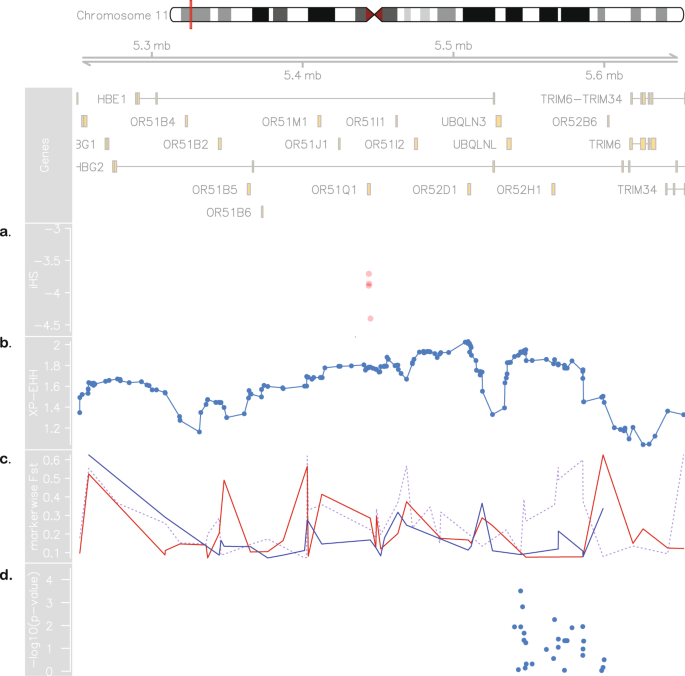
Evidence for positive pick in Brazilian sickle prison cell patients. At the top: chromosome 11 ideogram highlighting the region from 5.2 to 5.7 Mb, followed by genomic context. (a) Brazilian iHS values (values below −two indicate positive selection). (b) XP-EHH betwixt sickle cell anaemia cohorts from Brazil and US (values higher up 2 are considered signals of selection in one population but not in the other). (c) Pairwise SBR-SUS (dotted royal line), SBR-IBS (cherry-red line) and SBR-LWK (blueish line) FST values. (d) Association between makers in the 5.45–5.59 Mb range and HbF levels in the Brazilian accomplice.
We found local FST values on the v.45–5.59 Mb range to be lower when comparing Iberian (IBS) and Eastern African Bantu (LWK) populations to SBR (Fig. 3c red and blue lines, respectively), suggesting that this region may nowadays an introgression from Iberian origin. The FST values in the Bantu/Iberian comparison to SBR are still fairly high (to a higher place 0.iii), consistent with a scenario of positive pick. The hypothesis of introgression is likewise corroborated by the European beginnings local estimates in this location (Supplementary Fig. S2). Nosotros then tested LD betwixt the markers presenting atypical iHS values and markers around the rs334 mutation (untyped) region to evaluate if the selection signal is a product of malaria resistance and establish no linkage betwixt the two regions (Supplementary Fig. S3). Nosotros also compared SBR to BRZ and found this region to have FST values every bit high as 0.76 thereby ruling out the hypothesis that this signal is derived from a selective pressure that all Brazilians undergo.
Due to the implication of this region in the product of gamma globin, we performed association assay in a SBR subset. In doing so, we found two linked SNPs to exist positively associated with HbF levels after correcting for historic period, sex and hydroxyurea treatment and adjusting p-values for multiple testing: rs1433567, p-value = 0.0096 and rs2010794, p-value = 0.046 (run into Figs 3d, S4 and Supplementary Table S3). The markers are located in the LCR region, in the olfactory receptor gene cluster upstream to HBB and have not been reported in clan with HbF before. Moreover, these markers are in LD with regions comprising BCL11A biding sites described in Liu et al.14 and RFLP sites used in the HBB haplotype assignment (Supplementary Fig. S3).
Discussion
In the present study, we compared SCA patients from the U.s.a. and Brazil through the analysis of population structure at two levels, by genome-wide assay and by further exploring the mutation-harbouring chromosome. At the genomic level, the cohorts showed substantial differences with respect to ancestry. Nosotros constitute the Brazilian cohort to be more than admixed (Fig. one and Table 1) and more than likely to have greater European and Amerindian ancestries, while the US sample has a more prominent African background. Brazilian bequeathed proportions agree with a previous report on a sickle cell disease sample analysed on the continental ancestries level8.
By subdividing ancestry origins further to the subcontinental calibration, the North American cohort had a pattern of within-Africa ancestry consistent with reports of genetic relatedness to Yorubans9,xv. In a large study, Tishkoff et al. examined 4 African American populations along with 181 global populations and concluded that the quondam take ancestry predominantly from West-Africa (approx. 71%), followed by Europe (approx. 13%), other African regions (approx. 8%) and America (approx. 4%)16. They also described the African Americans to have a 45% Bantu mean ancestry and 22% non-Bantu (Mandinka ethnolinguistic grouping) mean ancestry, emphasizing that the diaspora encompassed a wide region in Africa, ranging from Senegambia in the w all the style to Republic of angola, in the southward16. Our data are consistent with these findings for both the SUS population and not-affected African descendants from the US, which are nearly identical in ancestral limerick.
Brazilian affected and unaffected subjects, on the other hand, are somewhat discernible by both PCA and ADMIXTURE plots, although we assert that the non-afflicted sample was non controlled for skin pigmentation and was rather nerveless at random. More than importantly, Brazilian HbAA were all collected in São Paulo, while the SCA group has too subjects from Pernambuco. Still, this differentiation is markedly small (FST = 0.001; Supplementary Table S2) and advocates for a higher admix rate in the Brazilian SCA cohort compared to the U.s.a. cohort analysed. The former has two-thirds of its African heritage traced to the East-African Bantu population, and the other i-tertiary to West-African non-Bantu populations. Although Brazilian predominance of Bantu limerick is consistent with reported migration records, the SUS group shows a internet contribution that is greater than what we observed for Brazilians. This might reverberate the Bantu expansion, one of the major demographic movements in history of mankind, thought to accept started around five m years ago, when Bantu-speaking people from Nigeria/Cameroon spread East and South, a migration probably prompted by agriculture17.
Unlike the SCA population from the US (meet Solovieff et al.ix), Brazilian SCA has only been briefly described in terms of genetic structure and ancestry8, and to the all-time of our knowledge, to appointment no subcontinental beginnings has ever been evaluated in this population. Kehdy et al. evaluated 6,487 subjects from the general populations of Northeast, Southeast and South Brazil, finding them to display two distinct within-Africa beginnings components: non-Bantu Western and Bantu Eastern and that the former was more prominent in Northeast Brazil, while the latter is more than prominent in the South-eastern/Southern areas. Withal, Bantu only accounted for an average of 36% in Southeastern people and 44% in Southern Brazilians, while nosotros plant Brazilians, irrespective of affliction condition, to share 65% of their African heritage traced to Bantu on average. This might exist due to the different regional origins of the recruited subjects and/or other methodological and belittling aspects, although both are in agreement with historiographical data, which states that enslaved Yoruban people arrived in large numbers in the Northeast port of Salvador, whereas the Mozambican Bantu slaves disembarked largely in Rio de Janeiro ports, in Due south-eastern Brazil18. Besides, Hudson'due south FST on genomic markers confirms that our sample of SCA from Brazil is slightly closer to Bantu than to non-Bantu populations. SCA individuals from the United states of america display a more even sub-continental African composition and greater proximity to the African populations evaluated here, indicating assortative mating may have had great impact on the Usa cohort. It is noteworthy that the FST values also show that the two affected cohorts are closer to each other than they are to European populations, and that the SBR accomplice is closer as well to its US counterpart than to any African population surveyed.
We establish that chromosome eleven haplotype ancestries in SCA cohorts generally correspond to the genome-wide ancestry proportions we establish in the previous analysis. Moreover, inferred HBB haplotypes agreed with the expected distribution: Machine prevails in SBR, while in SUS the Benin haplotype predominates. The HBB haplotypes were firstly believed to indicate five distinct HbS mutation events, only a contempo study favours the hypothesis of a unmarried origin of the HbS allele in Africa approximately 7,300 years agone19, while another written report, taking population construction, census, overdominance and balanced selection into account, estimated the origin of HbS mutation to accept taken place approximately 22,000 years agone in the ancestors of African agriculturalists20. By evaluating 20 haplotypes containing the HbS in the 1,000 Genomes Project and in Qatar subjects, Shriner and Rotimi identified three clusters resulting from two dissever events. The kickoff occurred on the ancestral haplotype and accounts for the Machine, Cameroon and Indian-Arab haplotypes, while the second gave ascent to two clusters, one accounting for the Senegal and the other accounting for the Benin haplotypes19. The authors proposed that HbS had a single origin in the Sahara or in West-Cardinal Africa, and a population diverged in nowadays-twenty-four hours Cameroon, carrying the first cluster east and southward as role of the Bantu expansion, while a split migration wave headed north and west to present-day Senegal and the Gambia, giving rise to the Senegal and Republic of benin haplotypes19.
Moreover, we propose that the deviation on the chromosome eleven is due to a recent choice event in the SBR population. We tested the genotyping charge per unit for this range and found no missing data for either population, and the proportions of HBB haplotypes are in understanding with those reported for both cohorts21. Option-suggestive signals seem to agree on a 100 kb region, as evaluated by LD, haplotype pattern, FST, iHS and XP-EHH (Figs 2 and 3), ranging from chromosome xi:5.iv Mb to 5.five Mb. This range comprises the LCR, a regulatory element well known for modulating the expression of gamma-globin. Low iHS values in the Brazilian patients overlap with a region too known to harbour an olfactory receptor cluster that has been associated with HbF production22. Additionally, we detected markers significantly associated with HbF in a group of 68 Brazilian patients subsequently correcting for age, sex activity and hydroxyurea treatment (Fig. 3d). This finding supports the hypothesis of a selection effect driven by an HbF modulating variant. Our data seem to exist consistent with those of the study by Creary et al.10, who reported an association between European ancestry and the proportion of erythrocytes containing HbF. Another study, from Leonardo et al. evaluated variants in 244 sickle cell patients and found rs9399137 in the HMIP-2 locus, a relatively common European polymorphism, significantly associated with HbF levels23. The relationship betwixt European background and clinical outcome is, therefore, far from established.
An alternative explanation for the local ancestry results is that the signals are a by-product of malaria related selection interim on the sickle cell allele. A hypothetical higher incidence of malaria in Brazil compared to the Us throughout history (malaria was controlled for well-nigh of the United states of america territory from the outset of the twentieth century on24) could influence LD patterns and generate the aforementioned results. We, and then, tested LD between the rs334 mutation region and the region nether selection and establish that they form independent blocks, not uncommonly linked at any marker (Supplementary Fig. S3). Moreover, FST values between affected and unaffected Brazilians are every bit loftier equally 0.76 in this region, implying that the putative selection result acts strictly on SCA subjects and is related to the disease and non to the full general population.
This study was express by the relatively small sample sizes in SCA cohorts derived from simply iii sampling localities. These limitations go far hard to extrapolate the results to larger and more than broadly distributed sickle prison cell individuals from the two countries evaluated and also dilate statistical racket. Although assessed in regard of IBD, individuals might however have cryptic structure/consanguinity that would especially affect the LD patterns observed for Brazilian patients. Differences in gene flow, HbS allele frequency and HBB haplotype composition between sampled subjects from Recife and Campinas may accept introduced variance not accounted in the assay. Although genotyping rate is near 100% for markers included in ancestral analysis (see Methods), technical constraints may utilize, equally the inference of haplotype phase by population data is known to accept greater switch error rates. Lo et al. evaluated major phasing algorithms and their accuracy through variation of panels and sample sizes, also as by comparing trio and populational phasing and institute SHAPEIT to yield a 3.52–6.51% switch error charge per unit in small unrelated datasets (N from 15 to 32)25, while Choi et al. plant SHAPEIT switch error to be 2.8% when phasing 85 unrelated individuals from European origin26. Nosotros would thus wait our information to fall into the range of approximately 3–6% switch error rate. The local ancestry inference might likewise be afflicted by the utilize of East Asian reference data as proxy of ancestral Americans, since information technology might inflate the estimates of haplotype contribution of that particular population27.
Here, we quantified divergence between two small-scale cohorts and constitute this to be a promising way to highlight regions of high divergence that might be of functional importance or to uncover candidate loci based on option signals. The haplotype construction has important implications on the cis-interim factors leading to variation on HbF product. More more often than not, these findings underline that the five RFLP-haplotype classifications proposed do not business relationship for population-specific demographic factors and, while nonetheless useful, should exist analysed advisedly.
Genetic studies struggle to deal with admixture and other complex population demographic characteristics in face up of clan to phenotypic traits. Admixture mapping, a tool to perform this task, has been recently adult and relies on regions of different allele frequency driven by contrasting ancestries. Information technology has been suggested that admixture mapping may only be applicable when bequeathed populations differ in the phenotype of interest8, and this seems to be the instance for SCA patients with regard to HbF production. Admixed mapping, nonetheless, has been applied when the ancestral populations are European and African10,28,29,30,31. It is however a affair of debate whether HbF levels are influenced by European ancestry8, whereas different ancestries inside the African continent take already been proven to be diverse regarding gamma-globin expression. Moreover, it is notwithstanding unclear how dissimilar levels of admixture volition interpret to HbF production and other phenotypic traits. Sickle cell affliction ancestry studies could lead to novel loci associated with phenotypic variability. Here nosotros demonstrate that SCA samples from different locations may largely vary on the genomic and local ancestry on chromosome 11. Further studies in larger cohorts, sampled from different locations are welcomed to amend draw the variation in ancestral background on genomic and HBB cluster levels. Besides, more detailed migration history data and the advancement in fine structure inference methods will broaden our understanding of how patterns of gene period, admixing, selection and linkage disequilibrium act on shaping genomic regions that touch on important phenotypic human traits.
In decision, we found the two different cohorts of SCA to differ in both genome-broad ancestral composition and locally to the causal locus region. Comparing admixed populations may be a strategy to reveal regions of local accommodation that would otherwise require a big clan study to be unveiled.
Cloth and Methods
Ethics statement
The present inquiry followed the principles of the Declaration of Helsinki; all patients were presented to the aims and details of the study and signed an informed consent. The institutional review board committee at CHOP and Ethics Committee at the Faculty of Medical Sciences at UNICAMP approved subjects' enrolment, blood collections and report methods.
Subjects
The complete dataset comprises a total of one,994 individuals, ane,822 of which are function of the 1,000 Genomes repository (http://www.internationalgenome.org/information/) representing global populations (Supplementary Table S1). Brazilian SCA patients were recruited at HEMOPE (Recife, Pernambuco; n = 57) and HEMOCENTRO (Campinas, São Paulo; n = 34) haematological therapy centres, along with 51 unaffected Brazilians from HEMOCENTRO (n = 31) and from the project "Cess of Copy Number Variation in Congenital Defects of Circuitous Inheritance"32, as well collected in Campinas (n = 20). The American cohort data is composed of 30 SCA patients with information filtered out from the Epic Intendance Clinical System (Ballsy, Verona, WI), along with 60 automobile-declared African Americans not affected by sickle cell diseases, all from CHOP, Pennsylvania. Differently from African-Americans, the Brazilian group of unaffected subjects (HbAA) was not selected regarding skin pigmentation or self-declared African groundwork, since the SBR population is already known to be more heterogeneous in ancestral limerickeight.
Genotyping
Genotyping of both American and Brazilian samples was carried out on the Affymetrix Genome-Wide Homo SNP 6.0 array platform (Affymetrix Inc., CA, USA), according to manufacturer's protocol. Genotype data was analysed along with reference populations from k Genomes Project33,34. Nosotros selected 19 reference populations from the African, European, American and Asian continents, as shown on Supplementary Table S1.
Quality command (QC)
Processing of raw genotype information and the bones quality control procedure was performed with the help of the PLINK v1.9 software35. Each private sample was checked for discordance in relation to the sex register, outlying missing genotype call rate (genotyping charge per unit ≥ 0.ninety were kept); nosotros as well evaluated relatedness in the collected samples past calculating genome-broad identity-by-descent (IBD), removing one sample from pairs of duplicates or pairs estimated to be 2nd-degree relatives or closer (IBD < 0.1875 were kept, see Supplementary Fig. S5). Each population was evaluated for genotyping quality and markers consistency throughout the sample, removing markers with low pocket-size allele frequency (<0.01); or demonstrating deviation from Hardy-Weinberg equilibrium (HWE), p-value < ten−9. We as well composed a list of SNPs for which at least one Mendelian inconsistency was observed in populations that had information for trios, excluding such SNPs from further analyses. The final genotyping telephone call rate was 0.9993 for genomic analysis and 1.0 and 0.9950 for chromosome 11 in SBR and SUS, respectively.
Linkage disequilibrium (LD)
For performing PCA, we accept controlled the information for regions of high LD through the extraction of local and long-range markers in LD (r2 < 0.5) as the first pace. We also excluded regions of known extensive LD beyond the genome36. For displaying regions of LD nosotros used Haploview v4.two37, SNPs with strong LD (D′ ≥0.viii) were considered function of a haplotype block using conviction intervals as proposed past Gabriel and colleagues38.
Genome-wide population construction
Genome-wide population structure and admixture were analysed by principal component analysis (PCA), using EIGENSOFT v7.2.139; and an beginnings modelling approach implemented in ADMIXTURE v1.3.040, while R software was used to generate graphical representation of the results. EIGENSOFT applied PCA, a non-parametric technique for reducing the multidimensionality to orthogonal eigenvectors that enclose the maximum variance to the genotypic data, in information from all 23 populations. Data is converted to a matrix representing individuals and their genotypes for 155,820 SNPs kept later on QC and LD treatment. Eigenvectors representative of the largest amount of variance in information were so used to build the PCA plot. We too calculated the F-statistic among populations, past the Hudson'due south FST method, also implemented in the EIGENSOFT package. ADMIXTURE software assigns individuals, on the basis of differences in allelic frequencies past maximum likelihood estimation, to beginnings clusters (K). We identified the optimal value of K (six) past the least error cantankerous-validation method afterward testing K values ranging from 1 to xviii.
Local ancestry inference
We phased genotypes of the chromosome eleven on both Brazilian and American SCA patients using the SHAPEIT v2.r790 method41. We then analysed local ancestry in this chromosome using the software SABER+ v1.0, which implements a Markov-Hidden Markov Model for inferring locus-specific ancestry in admixed individuals42. We modelled SCA cohorts as a mixture of chromosomes from three ancestral populations with diverse global proportions of European, Native American and Due west African ancestries. Although real admixture histories are more complex than this, nosotros simplified them for the sake of data tractability, since more convoluted admixing models are still poorly addressed by current algorithms43.
We considered admixed haplotypes every bit mosaics of segments derived from iii of the HapMap phase 3 haplotype panels34: phased haplotypes from the CEU (117 haplotypes), CHB + JPT (169) and YRI (115) trio-phased panels, every bit proxy haplotype data from Europeans, Native American and African ancestors, respectively. We too applied a Weir & Cockerham's makerwise FST estimation44 implemented in VCFLIB package (https://github.com/vcflib/vcflib). Haplotypes blocks images were generated in Haploview37 and VCFLIB.
HBB haplotypes inference
For evaluating inference in the classical 5 HBB haplotypes we used phased haplotypes of SCA subjects to impute not-typed markers on chromosome 11, including 4 SNPs (rs3834466, rs28440105, rs10128556, and rs968857) that ascertain these haplotypes as described in Shaikho et al.45. Imputation was performed by IMPUTE2 v2.3.2 method46.
Integrated haplotype score (iHS)
The integrated haplotype Score (iHS) was proposed by Voight et al. as a method to describe events of recent pick47. iHS is the amount of extended haplotype homozygosity (EHH) at a given marker along the ancestral allele relative to the derived allele empirically standardized to mean of 0 and variance of 1. Values lower than −ii (for ancestral allele) or higher than ii (for derived allele) are regarded as signals of recent positive selection. A stretch of extended homozygosity for haplotypes on a high frequency allele relative to the other is a signature of a sweep resulting from positive selection. We computed iHS values for SCA from Brazil and the US using the VCFLIB package. Past linearly interpolating between SNPs, EHH was integrated with respect to genetic distance for markers that reached EHH of 0.05 in both directions from the core SNP, otherwise, that SNP was skipped. Normalization is so performed to business relationship for regional differences in allele frequencies.
Cross-population extended homozygosity (XP-EHH)
Cantankerous Population Extended Haplotype Homozygosity detects sweeps resulting from selected alleles that have trend towards fixation in i population only non the other48. We used the selscan v1.1.0 software to perform XP-EHH calculation49.
Association exam
We selected the GRCh37 chr11:5.54–5.59 Mb region on account of the FST values shown in Fig. 3c. In this region, FST values are high between Brazilian and American cohorts merely drop between Brazilian and Iberian populations. We modelled HbF as response variable on a linear regression, performing a single test for each of the 31 SNPs as predictor variables, along with historic period, sexual activity and hydroxyurea treatment every bit covariates. All variants had MAF >0.05 and association with HbF was tested using a standard linear regression of phenotype on allele dosage implemented in PLINK v1.ix35. The gvlma R packagefifty was used to test fitness of data to the linear regression assumptions. We provide plots showing normality of residuals for significant SNPs in Supplementary Fig. S6. A significance level of 0.05 was adopted and Bonferroni aligning applied for correcting for multiple testing, We generated a local clan plot using the LocusZoom v1.451 tool (see Supplementary Fig. S4).
Data Availability
The datasets generated and/or analysed during the current study are bachelor from the corresponding author on reasonable request.
References
-
Herrick, J. Peculiar elongated and sickle-shaped red claret corpuscles in a example of severe anemia. Curvation. Intern. Med. xv, 490–493 (1910).
-
Rees, D. C., Williams, T. North. & Gladwin, Thousand. T. Sickle-cell disease. Lancet 376, 2018–31 (2010).
-
Loggetto, S. R. Sickle cell anemia: clinical diversity and beta S-globin haplotypes. Rev. Bras. Hematol. Hemoter. 35, 155–vii (2013).
-
Steinberg, 1000. H. & Sebastiani, P. Genetic modifiers of sickle prison cell disease. Am. J. Hematol. 87, 795–803 (2012).
-
Alsultan, A. et al. Sickle jail cell disease in Saudi Arabia: The phenotype in adults with the Arab-Indian haplotype is not benign. Br. J. Haematol. 164, 597–604 (2014).
-
Webster, G. T., Clegg, J. B. & Harding, R. G. Common 5′ beta-globin RFLP haplotypes harbour a surprising level of ancestral sequence mosaicism. Hum. Genet. 113, 123–39 (2003).
-
Liu, L. et al. High-density SNP genotyping to ascertain β-globin locus haplotypes. Blood Cells, Mol. Dis. 42, 16–24 (2009).
-
da Silva, M. C. F. et al. Extensive admixture in Brazilian sickle cell patients: implications for the mapping of genetic modifiers. Blood 118(4493–5), writer reply 4495 (2011).
-
Solovieff, N. et al. Ancestry of African Americans with sickle cell illness. Blood Cells. Mol. Dis. 47, 41–5 (2011).
-
Creary, Fifty. Due east. et al. Ethnic differences in F cell levels in Jamaica: a potential tool for identifying new genetic loci controlling fetal haemoglobin. Br. J. Haematol. 144, 954–60 (2009).
-
Piel, F. B., Hay, S. I., Gupta, S., Weatherall, D. J. & Williams, T. North. Global Burden of Sickle Cell Anaemia in Children nether Five, 2010–2050: Modelling Based on Demographics, Excess Bloodshed, and Interventions. PLoS Med. 10, e1001484 (2013).
-
Giolo, S. R. et al. Brazilian urban population genetic structure reveals a high degree of admixture. Eur. J. Hum. Genet. 20, 111–6 (2012).
-
Santos, H. C. et al. A minimum set of ancestry informative markers for determining admixture proportions in a mixed American population: the Brazilian set. Eur. J. Hum. Genet. ane–7, https://doi.org/10.1038/ejhg.2015.187 (2015).
-
Liu, N. et al. Straight Promoter Repression by BCL11A Controls the Fetal to Developed Hemoglobin Switch. Prison cell 173, 430–442.e17 (2018).
-
Montinaro, F. et al. Unravelling the subconscious ancestry of American admixed populations. Nat. Commun. 6, 1–vii (2015).
-
Tishkoff, Southward. A. et al. The Genetic Structure and History of Africans and African Americans. Science (80-.). 324, 1035–1044 (2009).
-
Berniell-Lee, G. et al. Genetic and demographic implications of the bantu expansion: Insights from human paternal lineages. Mol. Biol. Evol. 26, 1581–1589 (2009).
-
Pena, S. D. J. (Sergio D. J.. Homo brasilis: aspectos genéticos, lingüísticos, históricos e socioantropológicos da formação do povo brasileiro. (FUNPEC-RP, 2002).
-
Shriner, D. & Rotimi, C. N. Whole-Genome-Sequence-Based Haplotypes Reveal Unmarried Origin of the Sickle Allele during the Holocene Wet Phase. Am. J. Hum. Genet. 102, 547–556 (2018).
-
Laval, One thousand. et al. Contempo Adaptive Conquering by African Rainforest Hunter-Gatherers of the Belatedly Pleistocene Sickle-Cell Mutation Suggests By Differences in Malaria Exposure. Am. J. Hum. Genet. 104, 553–561 (2019).
-
Hattori, Y., Kutlar, F., Kutlar, A., McKie, V. C. & Huisman, T. H. Haplotypes of beta Due south chromosomes amongst patients with sickle prison cell anemia from Georgia. Hemoglobin 10, 623–42 (1986).
-
Solovieff, N. et al. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5′ olfactory receptor factor cluster. Claret 115, 1815–22 (2010).
-
Leonardo, F. C. et al. Reduced charge per unit of sickle-related complications in Brazilian patients carrying HbF-promoting alleles at the BCL11A and HMIP-two loci. Br. J. Haematol. 173, 456–460 (2016).
-
Hay, S. I., Guerra, C. A., Tatem, A. J., Noor, A. 1000. & Snow, R. Westward. The global distribution and population at hazard of malaria: past, present, and futurity. Lancet. Infect. Dis. 4, 327–36 (2004).
-
Loh, P. et al. Technical reports Reference-based phasing using the Haplotype Reference Consortium panel. 48 (2016).
-
Choi, Y., Chan, A. P., Kirkness, E., Telenti, A. & Schork, Due north. J. Comparison of phasing strategies for whole man genomes. PLoS Genet. 14, ane–26 (2018).
-
Baran, Y. et al. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics 28, 1359–1367 (2012).
-
Winkler, Ca, Nelson, M. W. & Smith, M. Due west. Admixture mapping comes of age. Annu. Rev. Genomics Hum. Genet. 11, 65–89 (2010).
-
Zhu, X., Tang, H. & Risch, N. Admixture Mapping and the Role of Population Structure for Localizing Disease Genes. Adv. Genet. threescore, 547–569 (2008).
-
Adler, S. et al. Mexican-American admixture mapping analyses for diabetic nephropathy in type 2 diabetes mellitus. Semin. Nephrol. 30, 141–149 (2010).
-
Reich, D. et al. A whole-genome admixture browse finds a candidate locus for multiple sclerosis susceptibility. Nat. Genet. 37, 1113–8 (2005).
-
Simioni, K., Araujo, T. K., Monlleo, I. 50., Maurer-Morelli, C. Five. & Gil-da-Silva-Lopes, 5. 50. Investigation of genetic factors underlying typical orofacial clefts: mutational screening and copy number variation. J. Hum. Genet. lx, 17–25 (2015).
-
Durbin, R. K. et al. A map of man genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
-
International, T. & Consortium, H. The International HapMap Project. Nature 426, 789–796 (2003).
-
Purcell, S. et al. PLINK: a tool fix for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–75 (2007).
-
Price, A. L. et al. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 83(132–v), author reply 135–9 (2008).
-
Barrett, J. C., Fry, B., Maller, J. & Daly, Thousand. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–5 (2005).
-
Gabriel, S. B. et al. The Structure of Haplotype Blocks in the Human Genome. Scientific discipline (80-.). 296, 2225–2229 (2002).
-
Patterson, N., Price, A. Fifty. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
-
Alexander, D. H., Novembre, J. & Lange, Yard. Fast model-based interpretation of ancestry in unrelated individuals. Genome Res. nineteen, 1655–1664 (2009).
-
Delaneau, O., Zagury, J.-F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods x, 5–six (2012).
-
Tang, H., Coram, M., Wang, P., Zhu, X. & Risch, North. Reconstructing genetic beginnings blocks in admixed individuals. Am. J. Hum. Genet. 79, 1–12 (2006).
-
Liu, Y. et al. Softwares and methods for estimating genetic ancestry in human populations. Hum. Genomics 7, 1 (2013).
-
Weir, B. South. & Cockerham, C. C. Estimating F-Statistics for the Analysis of Population Construction. Evolution (N. Y). 38, 1358 (1984).
-
Shaikho, E. M. et al. A phased SNP-based nomenclature of sickle prison cell anemia HBB haplotypes. 1–seven, https://doi.org/ten.1186/s12864-017-4013-y (2017).
-
Howie, B. N., Donnelly, P. & Marchini, J. A Flexible and Accurate Genotype Imputation Method for the Side by side Generation of Genome-Wide Clan Studies. PLoS Genet. five, e1000529 (2009).
-
Voight, B. F., Kudaravalli, S., Wen, X. & Pritchard, J. M. A map of recent positive selection in the human genome. PLoS Biol. 4, 0446–0458 (2006).
-
Sabeti, P. C. et al. Genome-wide detection and label of positive selection in human populations. Nature 449, 913–918 (2007).
-
Szpiech, Z. A. & Hernandez, R. D. Selscan: An Efficient Multithreaded Program to Perform EHH-Based Scans for Positive Choice. Mol. Biol. Evol. 31, 2824–2827 (2014).
-
Peña, East. A. & Slate, E. H. Global Validation of Linear Model Assumptions. J. Am. Stat. Assoc. 101, 341 (2006).
-
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association browse results. Bioinformatics 26, 2336–2337 (2010).
Acknowledgements
We would like to acknowledge grants from São Paulo Research Foundation (2008/57441-0, 2014/00984-3 - F.F.C.; 2012/06438-5, 2015/13152-9 - P.R.South.C.; and 2008/10596-0 - V.L.M.S.L) and Coordination for the Improvement of Higher Education Personnel/Council of Technological and Scientific Development (8367/2011-1, 150398/2013-one - G.A.; 304455/2012-1 - V.Fifty.G.S.L; and 310938/2014-7, 305218/2017-4 - M.B.M.). The Brazilian Synchrotron Light Laboratory and the Centre for Practical Genomics at the Childrens' Hospital of Philadelphia also supported this work.
Writer information
Affiliations
Contributions
P.R.Due south.C., Thou.A. and Yard.B.Thou. designed the study. P.R.South.C., G.A., M.Southward., I.F.D. and F.1000. conducted the experiments, A.S.A., H.H. and F.F.C. enabled access to patients and assisted with clinical information, F.F.C., V.L.G.S.L., A.S.A., G.A.C.B., I.F.D., R.P., H.H. and 1000.A. contributed recruiting individuals, providing clinical and demographical information and gathering array data, P.R.South.C. and G.A. conducted data assay. One thousand.B.M., F.F.C. and H.H. obtained resources. P.R.Due south.C. wrote the manuscript. All authors reviewed and canonical the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Boosted information
Publisher'due south note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Eatables Attribution 4.0 International License, which permits employ, sharing, adaptation, distribution and reproduction in any medium or format, equally long as you give appropriate credit to the original author(southward) and the source, provide a link to the Creative Commons license, and point if changes were made. The images or other third party cloth in this commodity are included in the commodity'south Creative Commons license, unless indicated otherwise in a credit line to the fabric. If fabric is not included in the article'south Creative Commons license and your intended use is not permitted past statutory regulation or exceeds the permitted utilise, you lot will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
Near this article
Cite this article
Cruz, P.R.S., Ananina, K., Gil-da-Silva-Lopes, V.50. et al. Genetic comparing of sickle jail cell anaemia cohorts from Brazil and the United states of america reveals high levels of deviation. Sci Rep nine, 10896 (2019). https://doi.org/10.1038/s41598-019-47313-2
-
Received:
-
Accepted:
-
Published:
-
DOI : https://doi.org/ten.1038/s41598-019-47313-two
Comments
By submitting a comment you lot concur to bide by our Terms and Customs Guidelines. If y'all observe something abusive or that does not comply with our terms or guidelines delight flag it as inappropriate.
Source: https://www.nature.com/articles/s41598-019-47313-2
0 Response to "sickle cell anemia has been traced to what type of mutation?"
Post a Comment