As Growing Cells Replicate the Two Replicated Dna Origins Split Again
Arthur Kornberg compared DNA to a tape recording of instructions that can be copied over and over. How do cells make these near-perfect copies, and does the process e'er vary?
Scientists take devoted decades of try to agreement how deoxyribonucleic acid (Dna) replicates itself. In uncomplicated terms, replication involves apply of an existing strand of Dna as a template for the synthesis of a new, identical strand. American enzymologist and Nobel Prize winner Arthur Kornberg compared this process to a tape recording of instructions for performing a task: "[East]xact copies tin can be fabricated from it, as from a tape recording, then that this data can exist used again and elsewhere in time and space" (Kornberg, 1960).
In reality, the process of replication is far more complex than suggested by Kornberg's analogy. Researchers typically utilize simple bacterial cells in their experiments, but they all the same exercise not have all the answers, particularly when it comes to eukaryotic replication. Nonetheless, scientists are familiar with the basic steps in the replication process, and they continue to rely on this information as the ground for continued inquiry and experimentation.
The Molecular Machinery of Bacterial Deoxyribonucleic acid Replication
A typical bacterial prison cell has anywhere from about ane 1000000 to 4 million base pairs of DNA, compared to the iii billion base pairs in the genome of the common house mouse (Mus muscle). Still, even in bacteria, with their smaller genomes, DNA replication involves an incredibly sophisticated, highly coordinated series of molecular events. These events are divided into 4 major stages: initiation, unwinding, primer synthesis, and elongation.
Initiation and Unwinding
During initiation, and then-called initiator proteins bind to the replication origin, a base of operations-pair sequence of nucleotides known as oriC. This binding triggers events that unwind the DNA double helix into 2 single-stranded Deoxyribonucleic acid molecules. Several groups of proteins are involved in this unwinding (Figure 1). For instance, the DNA helicases are responsible for breaking the hydrogen bonds that join the complementary nucleotide bases to each other; these hydrogen bonds are an essential feature of James Watson and Francis Crick's three-dimensional Dna model. Because the newly unwound single strands have a tendency to rejoin, another grouping of proteins, the single-strand-binding proteins, go on the unmarried strands stable until elongation begins. A third family unit of proteins, the topoisomerases, reduce some of the torsional strain caused by the unwinding of the double helix.
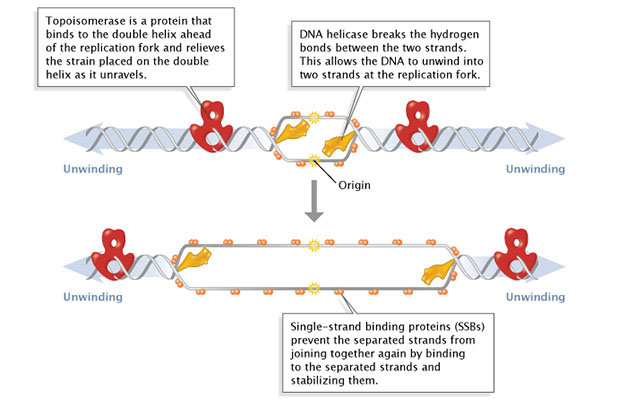
Effigy 1: Facilitation of Dna unwinding.
During DNA replication, several proteins facilitate the unwinding of the Deoxyribonucleic acid double helix into two single strands. Topoisomerases (red) reduce torsional strain caused by the unwinding of the Dna double helix; DNA helicase (yellow) breaks hydrogen bonds betwixt complementary base of operations-pairs; single-strand binding proteins (SSBs) stabilize the separated strands and prevent them from rejoining.
© 2014 Nature Education Adjusted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2d ed. All rights reserved. ![]()
As previously mentioned, the location at which a Deoxyribonucleic acid strand begins to unwind into ii separate single strands is known as the origin of replication. As shown in Figure one, when the double helix unwinds, replication proceeds along the two unmarried strands at the same fourth dimension but in opposite directions (i.eastward., left to right on 1 strand, and right to left on the other). This forms 2 replication forks that movement forth the DNA, replicating as they go.
Primer Synthesis
Primer synthesis marks the beginning of the actual synthesis of the new Deoxyribonucleic acid molecule. Primers are short stretches of nucleotides (about x to 12 bases in length) synthesized by an RNA polymerase enzyme called primase. Primers are required because DNA polymerases, the enzymes responsible for the actual addition of nucleotides to the new Deoxyribonucleic acid strand, can only add deoxyribonucleotides to the three'-OH group of an existing chain and cannot begin synthesis de novo. Primase, on the other hand, can add together ribonucleotides de novo. Later, after elongation is complete, the primer is removed and replaced with Deoxyribonucleic acid nucleotides.
Elongation
Finally, elongation--the addition of nucleotides to the new Deoxyribonucleic acid strand--begins after the primer has been added. Synthesis of the growing strand involves adding nucleotides, i by one, in the verbal order specified by the original (template) strand. Recall that one of the key features of the Watson-Crick Deoxyribonucleic acid model is that adenine is always paired with thymine and cytosine is ever paired with guanine. So, for example, if the original strand reads A-G-C-T, the new strand will read T-C-M-A.
DNA is e'er synthesized in the five'-to-3' direction, meaning that nucleotides are added only to the iii' end of the growing strand. Equally shown in Effigy 2, the v'-phosphate group of the new nucleotide binds to the 3'-OH group of the last nucleotide of the growing strand. Scientists have notwithstanding to identify a polymerase that tin can add bases to the 5' ends of Dna strands.
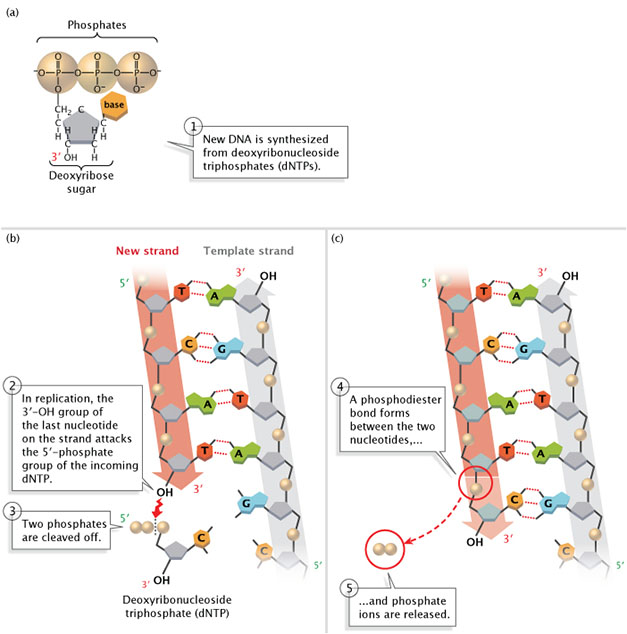
Effigy 2: New DNA is synthesized from deoxyribonucleoside triphosphates (dNTPs).
(A) A deoxyribonucleoside triphosphate (dNTP). (B) During DNA replication, the 3'-OH group of the concluding nucleotide on the new strand attacks the v'-phosphate group of the incoming dNTP. Two phosphates are cleaved off. (C) A phosphodiester bond forms between the 2 nucleotides, and phosphate ions are released.
© 2014 Nature Education Adapted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2nd ed. All rights reserved. ![]()
The Discovery of DNA Polymerase
While studying Due east. coli bacteria, enzymologist Arthur Kornberg discovered that Deoxyribonucleic acid polymerases catalyze Dna synthesis. Kornberg'southward experiment involved mixing all of the basic "ingredients" necessary for East. coli Dna synthesis in a test tube, including nucleotides, East. coli excerpt, and ATP, and so purifying and testing the enzymes involved. Using this method, Kornberg not only discovered Dna polymerases, merely he also performed some of the initial work demonstrating how enzymes add new nucleotides to growing DNA chains (Kornberg, 1959).
Scientists have since identified a total of five different DNA polymerases in E. coli, each with a specialized role. For case, Deoxyribonucleic acid polymerase III does nearly of the elongation piece of work, calculation nucleotides one by 1 to the iii' stop of the new and growing single strand. Other enzymes, including DNA polymerase I and RNase H, are responsible for removing the RNA primer after DNA polymerase Three has begun its work, replacing it with DNA nucleotides (Ogawa & Okazaki, 1984). When these enzymes finish, they get out a nick between the department of Dna that was formerly the primer and the elongated section of DNA. Another enzyme called Dna ligase then acts to seal the bond between the two next nucleotides.
Dna Polymerase But Moves in Ane Management
Afterward a primer is synthesized on a strand of DNA and the DNA strands unwind, synthesis and elongation can go along in only one direction. As previously mentioned, Dna polymerase tin can but add together to the 3' end, then the five' end of the primer remains unaltered. Consequently, synthesis gain immediately only along the so-chosen leading strand. This immediate replication is known as continuous replication. The other strand (in the 5' direction from the primer) is chosen the lagging strand, and replication forth information technology is chosen discontinuous replication. The double helix has to unwind a scrap earlier the synthesis of another primer can be initiated farther up on the lagging strand. Synthesis tin so occur from the 3' end of that new primer. Adjacent, the double helix unwinds a bit more, and another spurt of replication proceeds. Every bit a result, replication forth the lagging strand can only proceed in short, discontinuous spurts (Figure 3).
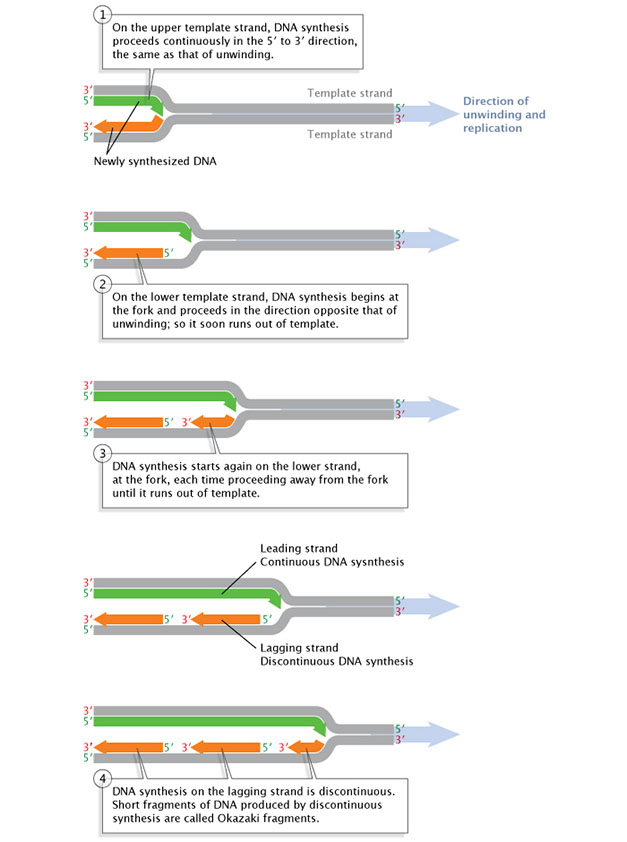
Effigy 3: Replication of the leading Dna strand is continuous, while replication along the lagging strand is discontinuous.
Later a short length of the DNA has been unwound, synthesis must proceed in the five' to three' direction; that is, in the direction opposite that of the unwinding.
© 2014 Nature Education Adapted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2nd ed. All rights reserved. ![]()
The fragments of newly synthesized Deoxyribonucleic acid forth the lagging strand are called Okazaki fragments, named in honour of their discoverer, Japanese molecular biologist Reiji Okazaki. Okazaki and his colleagues made their discovery past conducting what is known equally a pulse-hunt experiment, which involved exposing replicating Deoxyribonucleic acid to a short "pulse" of isotope-labeled nucleotides then varying the length of time that the cells would be exposed to nonlabeled nucleotides. This afterwards period is called the "chase" (Okazaki et al., 1968). The labeled nucleotides were incorporated into growing Dna molecules only during the initial few seconds of the pulse; thereafter, only nonlabeled nucleotides were incorporated during the chase. The scientists then centrifuged the newly synthesized DNA and observed that the shorter chases resulted in most of the radioactive decay appearing in "deadening" Dna. The sedimentation rate was determined by size: smaller fragments precipitated more slowly than larger fragments because of their lighter weight. As the investigators increased the length of the chases, radioactive decay in the "fast" Dna increased with little or no increase of radioactivity in the wearisome Dna. The researchers correctly interpreted these observations to hateful that, with short chases, but very small fragments of Deoxyribonucleic acid were being synthesized along the lagging strand. Every bit the chases increased in length, giving Dna more fourth dimension to replicate, the lagging strand fragments started integrating into longer, heavier, more than chop-chop sedimenting DNA strands. Today, scientists know that the Okazaki fragments of bacterial DNA are typically between 1,000 and 2,000 nucleotides long, whereas in eukaryotic cells, they are just about 100 to 200 nucleotides long.
The Challenges of Eukaryotic Replication
Bacterial and eukaryotic cells share many of the same basic features of replication; for instance, initiation requires a primer, elongation is always in the v'-to-iii' direction, and replication is ever continuous along the leading strand and discontinuous along the lagging strand. Only there are also important differences between bacterial and eukaryotic replication, some of which biologists are still actively researching in an effort to amend understand the molecular details. One difference is that eukaryotic replication is characterized by many replication origins (ofttimes thousands), not just one, and the sequences of the replication origins vary widely among species. On the other hand, while the replication origins for leaner, oriC, vary in length (from about 200 to 1,000 base pairs) and sequence, except among closely related organisms, all bacteria nonetheless have just a single replication origin (Mackiewicz et al., 2004).
Eukaryotic replication also utilizes a different set of DNA polymerase enzymes (e.thou., DNA polymerase δ and DNA polymerase ε instead of DNA polymerase Three). Scientists are yet studying the roles of the 13 eukaryotic polymerases discovered to date. In addition, in eukaryotes, the DNA template is compacted past the mode it winds around proteins chosen histones. This DNA-histone complex, called a nucleosome, poses a unique challenge both for the jail cell and for scientists investigating the molecular details of eukaryotic replication. What happens to nucleosomes during Dna replication? Scientists know from electron micrograph studies that nucleosome reassembly happens very chop-chop after replication (the reassembled nucleosomes are visible in the electron micrograph images), but they still practise not know how this happens (Annunziato, 2005).
Too, whereas bacterial chromosomes are circular, eukaryotic chromosomes are linear. During circular DNA replication, the excised primer is readily replaced by nucleotides, leaving no gap in the newly synthesized Deoxyribonucleic acid. In dissimilarity, in linear Deoxyribonucleic acid replication, at that place is always a small gap left at the very end of the chromosome considering of the lack of a iii'-OH group for replacement nucleotides to bind. (Equally mentioned, Dna synthesis can keep only in the 5'-to-3' direction.) If there were no style to fill this gap, the DNA molecule would become shorter and shorter with every generation. However, the ends of linear chromosomes—the telomeres—take several properties that prevent this.
DNA replication occurs during the S phase of jail cell division. In E. coli, this means that the entire genome is replicated in merely 40 minutes, at a footstep of approximately 1,000 nucleotides per second. In eukaryotes, the footstep is much slower: most 40 nucleotides per second. The coordination of the protein complexes required for the steps of replication and the speed at which replication must occur in order for cells to carve up are impressive, especially because that enzymes are also proofreading, which leaves very few errors backside.
Summary
The study of DNA replication started well-nigh as soon equally the structure of DNA was elucidated, and it continues to this day. Currently, the stages of initiation, unwinding, primer synthesis, and elongation are understood in the most basic sense, just many questions remain unanswered, particularly when it comes to replication of the eukaryotic genome. Scientists have devoted decades to the report of replication, and researchers such as Kornberg and Okazaki take fabricated a number of important breakthroughs. Still, much remains to be learned about replication, including how errors in this process contribute to human disease.
References and Recommended Reading
Annunziato, A. T. Split decision: What happens to nucleosomes during DNA replication? Journal of Biological Chemistry 280, 12065–12068 (2005)
Bessman, Yard. J., et al. Enzymatic synthesis of deoxyribonucleic acid. II. General properties of the reaction. Journal of Biological Chemistry 233, 171–177 (1958)
Kornberg, A. The biological synthesis of deoxyribonucleic acid. Nobel Lecture, December eleven, 1959. (link to transcript)
———. Biological synthesis of deoxyribonucleic acid. Science 131, 1503–1508 (1960)
Lehman, I. R., et al. Enzymatic synthesis of deoxyribonucleic acrid. I. Training of substrates and fractional purification of an enzyme from Escherichia coli. Periodical of Biological Chemistry 233, 163–170 (1958)
Losick, R., & Shapiro, L. DNA replication: Bringing the mountain to Mohammed. Science 282, 1430–1431 (1998)
Mackiewicz, P., et al. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Research 32, 3781–3791 (2004)
Ogawa, T., & Okazaki, T. Role of RNase H in Dna replication revealed past RNase H defective mutants of Escherichia coli. Molecular and General Genetics 193, 231–237 (1984)
Okazaki, R., et al. Machinery of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proceedings of the National Academy of Sciences 59, 598–605 (1968)
Source: http://www.nature.com/scitable/topicpage/major-molecular-events-of-dna-replication-413
0 Response to "As Growing Cells Replicate the Two Replicated Dna Origins Split Again"
Post a Comment